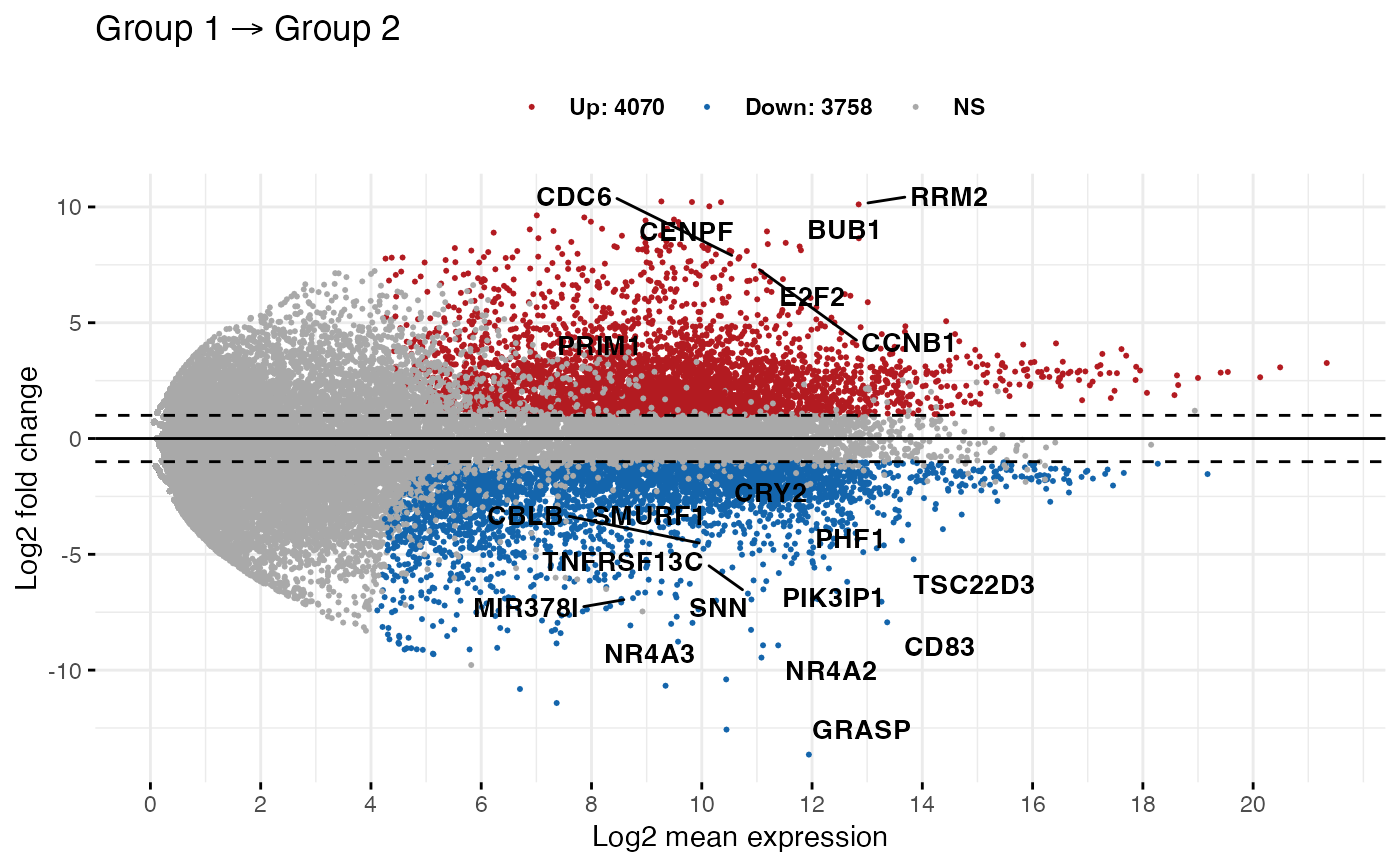
MD plot showing the log-fold change and average abundance of each gene.... | Download Scientific Diagram
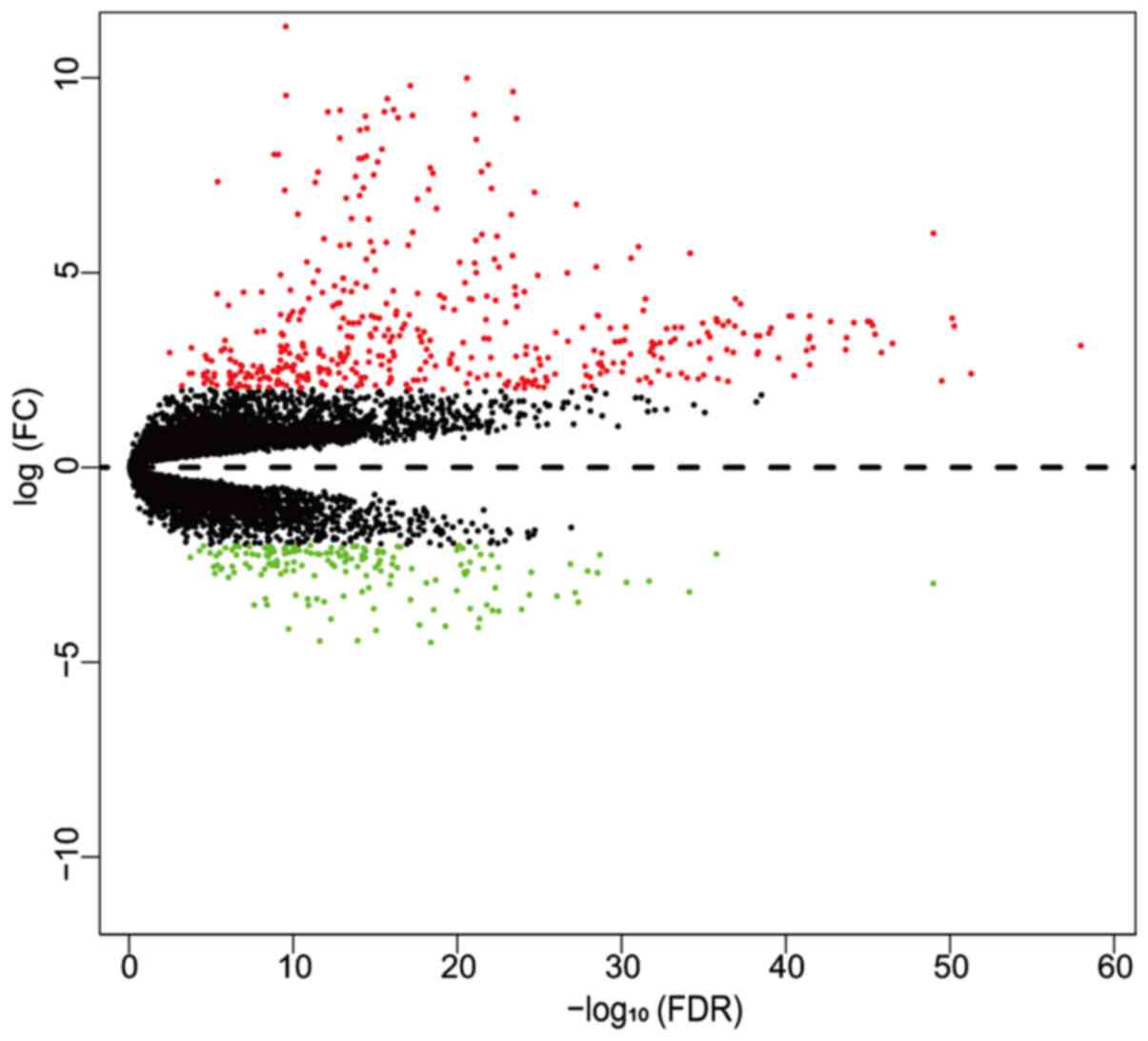
Identification of key genes and pathways by bioinformatics analysis with TCGA RNA sequencing data in hepatocellular carcinoma

Advantages of RNA‐seq compared to RNA microarrays for transcriptome profiling of anterior cruciate ligament tears - Rai - 2018 - Journal of Orthopaedic Research - Wiley Online Library

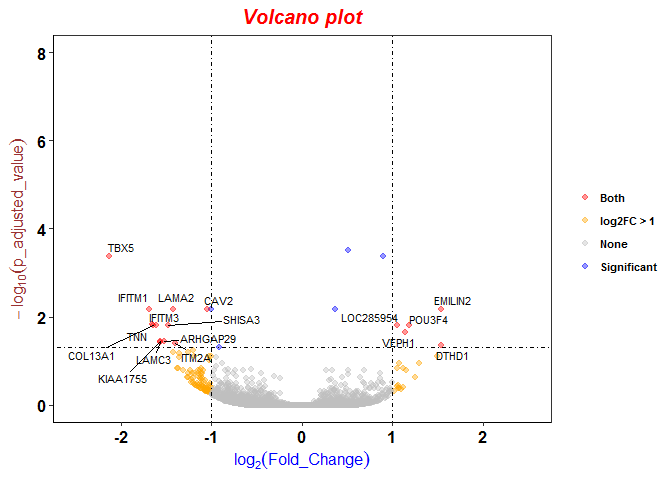
Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram
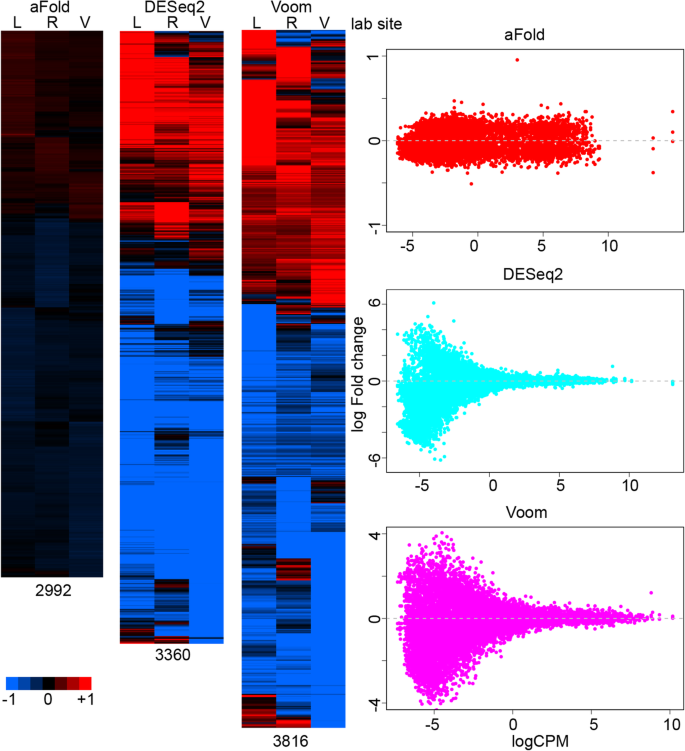
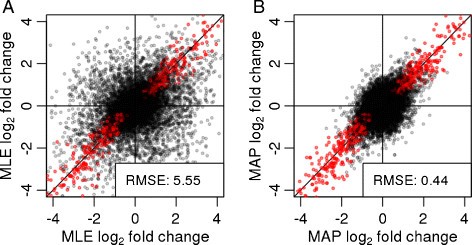
aFold – using polynomial uncertainty modelling for differential gene expression estimation from RNA sequencing data | BMC Genomics | Full Text
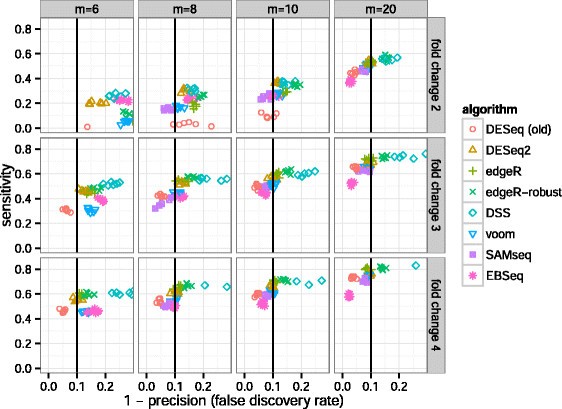
A comparison of per sample global scaling and per gene normalization methods for differential expression analysis of RNA-seq data

Tuning Transcriptional Regulation through Signaling: A Predictive Theory of Allosteric Induction - ScienceDirect

Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram
PLOS ONE: The hepatic transcriptome of the turkey poult (Meleagris gallopavo) is minimally altered by high inorganic dietary selenium

Differentially expressed protein groups (q < 0.4 and Log 2 fold change... | Download Scientific Diagram